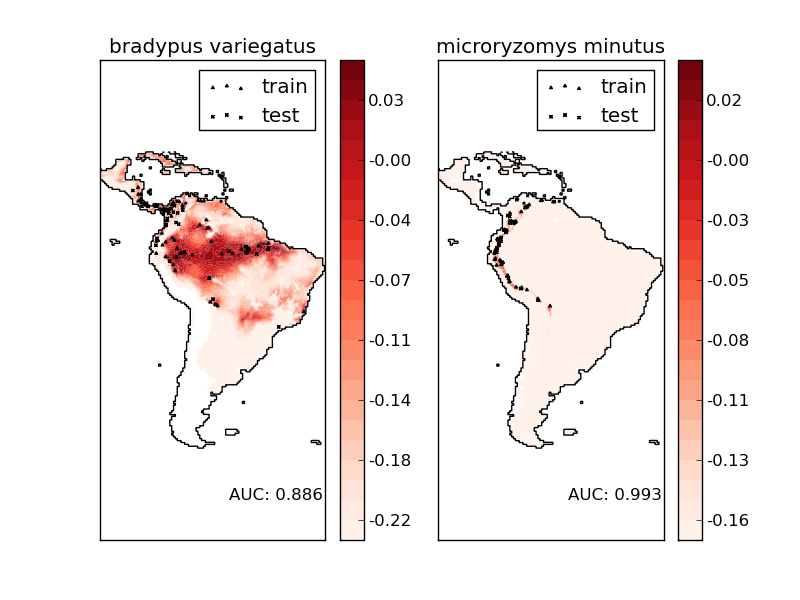
Species distribution modeling¶
Modeling species’ geographic distributions is an important problem in conservation biology. In this example we model the geographic distribution of two south american mammals given past observations and 14 environmental variables. Since we have only positive examples (there are no unsuccessful observations), we cast this problem as a density estimation problem and use the OneClassSVM provided by the package sklearn.svm as our modeling tool. The dataset is provided by Phillips et. al. (2006). If available, the example uses basemap to plot the coast lines and national boundaries of South America.
The two species are:
- Bradypus variegatus , the Brown-throated Sloth.
- Microryzomys minutus , also known as the Forest Small Rice Rat, a rodent that lives in Peru, Colombia, Ecuador, Peru, and Venezuela.
References:
- “Maximum entropy modeling of species geographic distributions” S. J. Phillips, R. P. Anderson, R. E. Schapire - Ecological Modelling, 190:231-259, 2006.
Python source code: plot_species_distribution_modeling.py
from __future__ import division
# Author: Peter Prettenhofer <peter.prettenhofer@gmail.com>
#
# License: Simplified BSD
print __doc__
import os
from os.path import normpath, split, exists
from glob import glob
from time import time
import pylab as pl
import numpy as np
try:
from mpl_toolkits.basemap import Basemap
basemap = True
except ImportError:
basemap = False
from sklearn import svm
from sklearn.metrics import roc_curve, auc
from sklearn.datasets.base import Bunch
###############################################################################
# Download the data, if not already on disk
samples_url = "http://www.cs.princeton.edu/~schapire/maxent/datasets/" \
"samples.zip"
coverage_url = "http://www.cs.princeton.edu/~schapire/maxent/datasets/" \
"coverages.zip"
samples_archive_name = "samples.zip"
coverage_archive_name = "coverages.zip"
def download(url, archive_name):
if not exists(archive_name[:-4]):
if not exists(archive_name):
import urllib
print "Downloading data, please wait ..."
print url
opener = urllib.urlopen(url)
open(archive_name, 'wb').write(opener.read())
print
import zipfile
print "Decompressiong the archive: " + archive_name
zipfile.ZipFile(archive_name).extractall()
# Remove the archive: we don't need it as we have expanded it
# to directory
os.unlink(archive_name)
print
download(samples_url, samples_archive_name)
download(coverage_url, coverage_archive_name)
t0 = time()
###############################################################################
# Define constants (coordinates, grid cells)
species = ["bradypus_variegatus_0", "microryzomys_minutus_0"]
species_map = dict([(s, i) for i, s in enumerate(species)])
# x,y coordinates of study area
x_left_lower_corner = -94.8
y_left_lower_corner = -56.05
# number of cells along x axis
n_cols = 1212
# number of cells along y axis
n_rows = 1592
grid_size = 0.05 # ~5.5 km
# x,y coordinates for corner cells
xmin = x_left_lower_corner + grid_size
xmax = xmin + (n_cols * grid_size)
ymin = y_left_lower_corner + grid_size
ymax = ymin + (n_rows * grid_size)
# x coordinates of the grid cells
xx = np.arange(xmin, xmax, grid_size)
# y coordinates of the grid cells
yy = np.arange(ymin, ymax, grid_size)
# The grid in x,y coordinates
X, Y = np.meshgrid(xx, yy[::-1])
print "Data grid"
print "---------"
print "xmin, xmax:", xmin, xmax
print "ymin, ymax:", ymin, ymax
print "grid size:", grid_size
print
###############################################################################
# Load data
def read_file(fname):
"""Read coverage grid data; returns array of
shape [n_rows, n_cols]. """
f = open(fname)
# Skip header
for i in range(6):
f.readline()
X = np.fromfile(f, dtype=np.float32, sep=" ", count=-1)
f.close()
return X.reshape((n_rows, n_cols))
def load_dir(directory):
"""Loads each of the coverage grids and returns a
tensor of shape [14, n_rows, n_cols].
"""
data = []
for fpath in glob("%s/*.asc" % normpath(directory)):
fname = split(fpath)[-1]
fname = fname[:fname.index(".")]
X = read_file(fpath)
data.append(X)
return np.array(data, dtype=np.float32)
print "loading data from disk..."
species2id = lambda s: species_map.get(s, -1)
train = np.loadtxt('samples/alltrain.csv', converters={0: species2id},
skiprows=1, delimiter=",")
test = np.loadtxt('samples/alltest.csv', converters={0: species2id},
skiprows=1, delimiter=",")
# Load env variable grids
coverage = load_dir("coverages")
# Per species data
bv = Bunch(name=" ".join(species[0].split("_")[:2]),
train=train[train[:, 0] == 0, 1:],
test=test[test[:, 0] == 0, 1:])
mm = Bunch(name=" ".join(species[1].split("_")[:2]),
train=train[train[:, 0] == 1, 1:],
test=test[test[:, 0] == 1, 1:])
def get_coverages(points, coverages, xx, yy):
"""Get coverages (aka features) for each point.
Returns
-------
array : shape = [points.shape[0], coverages.shape[0]]
The feature vectors (coverages) for each data point.
"""
rows = []
cols = []
for n in range(points.shape[0]):
i = np.searchsorted(xx, points[n, 0])
j = np.searchsorted(yy, points[n, 1])
rows.append(-j)
cols.append(i)
return coverages[:, rows, cols].T
# Get feature vectors (=coverages)
bv.train_cover = get_coverages(bv.train, coverage, xx, yy)
bv.test_cover = get_coverages(bv.test, coverage, xx, yy)
mm.train_cover = get_coverages(mm.train, coverage, xx, yy)
mm.test_cover = get_coverages(mm.test, coverage, xx, yy)
# background points (grid coordinates) for evaluation
np.random.seed(13)
background_points = np.c_[np.random.randint(low=0, high=n_rows, size=10000),
np.random.randint(low=0, high=n_cols, size=10000)].T
###############################################################################
# Helper functions
def predict(clf, mean, std):
"""Predict the density for each cell under model `clf`.
Only evaluates `clf` on land grid cells.
Note: Sea grid cells have coverage[2] equals -9999.
Returns
-------
array : shape [n_rows, n_cols]
A grid of same shape as X and Y which gives the density.
"""
Z = np.ones((n_rows, n_cols), dtype=np.float64)
# the land points
idx = np.where(coverage[2] > -9999)
X = coverage[:, idx[0], idx[1]].T
pred = clf.decision_function((X - mean) / std)[:, 0]
Z *= pred.min()
Z[idx[0], idx[1]] = pred
return Z
###############################################################################
# Fit, predict, and plot for each species.
for i, species in enumerate([bv, mm]):
print "_" * 80
print "Modeling distribution of species '%s'" % species.name
print
# Standardize features
mean = species.train_cover.mean(axis=0)
std = species.train_cover.std(axis=0)
train_cover_std = (species.train_cover - mean) / std
# Fit OneClassSVM
print "fit OneClassSVM ... ",
clf = svm.OneClassSVM(nu=0.1, kernel="rbf", gamma=0.5)
clf.fit(train_cover_std)
print "done. "
# Plot map of South America
pl.subplot(1, 2, i + 1)
if basemap:
print "plot coastlines using basemap"
m = Basemap(projection='cyl', llcrnrlat=ymin,
urcrnrlat=ymax, llcrnrlon=xmin,
urcrnrlon=xmax, resolution='c')
m.drawcoastlines()
m.drawcountries()
#m.drawrivers()
else:
print "plot coastlines from coverage"
CS = pl.contour(X, Y, coverage[2, :, :], levels=[-9999], colors="k",
linestyles="solid")
pl.xticks([])
pl.yticks([])
print "predict species distribution"
Z = predict(clf, mean, std)
levels = np.linspace(Z.min(), Z.max(), 25)
Z[coverage[2, :, :] == -9999] = -9999
CS = pl.contourf(X, Y, Z, levels=levels, cmap=pl.cm.Reds)
pl.colorbar(format='%.2f')
pl.scatter(species.train[:, 0], species.train[:, 1], s=2 ** 2, c='black',
marker='^', label='train')
pl.scatter(species.test[:, 0], species.test[:, 1], s=2 ** 2, c='black',
marker='x', label='test')
pl.legend()
pl.title(species.name)
pl.axis('equal')
# Compute AUC w.r.t. background points
pred_background = Z[background_points[0], background_points[1]]
pred_test = clf.decision_function((species.test_cover - mean) / std)[:, 0]
scores = np.r_[pred_test, pred_background]
y = np.r_[np.ones(pred_test.shape), np.zeros(pred_background.shape)]
fpr, tpr, thresholds = roc_curve(y, scores)
roc_auc = auc(fpr, tpr)
pl.text(-35, -70, "AUC: %.3f" % roc_auc, ha="right")
print "Area under the ROC curve : %f" % roc_auc
print "time elapsed: %.3fs" % (time() - t0)
pl.show()
