Kernel PCA¶
This example shows that Kernel PCA is able to find a projection of the data that makes data linearly separable.
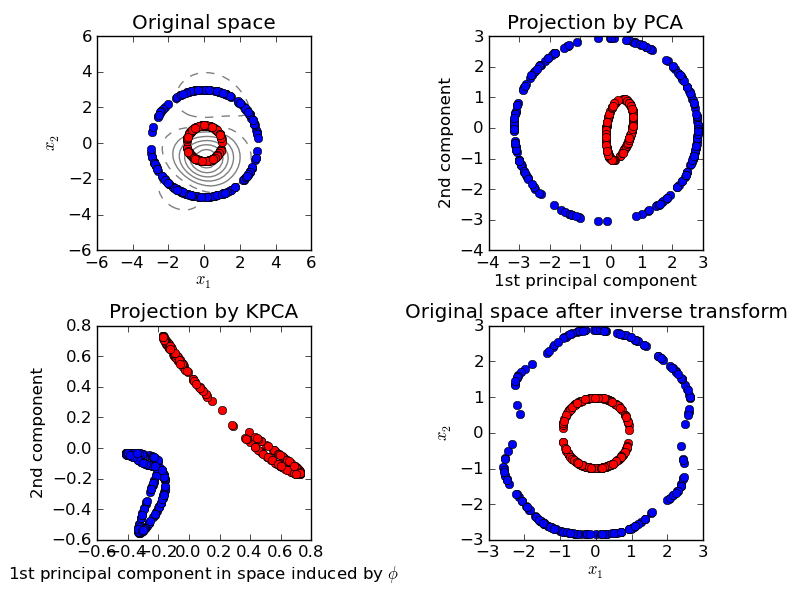
Python source code: plot_kernel_pca.py
print __doc__
# Authors: Mathieu Blondel
# License: BSD
import numpy as np
import pylab as pl
from sklearn.decomposition import PCA, KernelPCA
np.random.seed(0)
def genenerate_rings(n_samples=200):
x_red = np.random.random((n_samples,)) * 2 - 1
signs_red = np.sign(np.random.random(x_red.shape) - 0.5)
y_red = np.sqrt(np.abs(x_red ** 2 - 1)) * signs_red
x_blue = np.random.random((n_samples,)) * 6 - 3
signs_blue = np.sign(np.random.random(x_blue.shape) - 0.5)
y_blue = np.sqrt(np.abs(x_blue ** 2 - 9)) * signs_blue
return np.hstack(([x_red, y_red], [x_blue, y_blue])).T
def generate_clusters(n_samples=200):
mean1 = np.array([0, 2])
mean2 = np.array([2, 0])
cov = np.array([[2.0, 1.0], [1.0, 2.0]])
X_red = np.random.multivariate_normal(mean1, cov, n_samples)
X_blue = np.random.multivariate_normal(mean2, cov, n_samples)
return np.vstack((X_red, X_blue))
X = genenerate_rings()
#X = generate_clusters()
kpca = KernelPCA(kernel="rbf", fit_inverse_transform=True, gamma=0.5)
X_kpca = kpca.fit_transform(X)
X_back = kpca.inverse_transform(X_kpca)
pca = PCA()
X_pca = pca.fit_transform(X)
# Plot results
pl.figure()
pl.subplot(2, 2, 1, aspect='equal')
pl.title("Original space")
pl.plot(X[:200, 0], X[:200, 1], "ro")
pl.plot(X[200:, 0], X[200:, 1], "bo")
pl.xlabel("$x_1$")
pl.ylabel("$x_2$")
X1, X2 = np.meshgrid(np.linspace(-6, 6, 50), np.linspace(-6, 6, 50))
X_grid = np.array([np.ravel(X1), np.ravel(X2)]).T
# projection on the first principal component (in the phi space)
Z_grid = kpca.transform(X_grid)[:, 0].reshape(X1.shape)
pl.contour(X1, X2, Z_grid, colors='grey', linewidths=1, origin='lower')
pl.subplot(2, 2, 2, aspect='equal')
pl.plot(X_kpca[:200, 0], X_pca[:200, 1], "ro")
pl.plot(X_pca[200:, 0], X_pca[200:, 1], "bo")
pl.title("Projection by PCA")
pl.xlabel("1st principal component")
pl.ylabel("2nd component")
pl.subplot(2, 2, 3, aspect='equal')
pl.plot(X_kpca[:200, 0], X_kpca[:200, 1], "ro")
pl.plot(X_kpca[200:, 0], X_kpca[200:, 1], "bo")
pl.title("Projection by KPCA")
pl.xlabel("1st principal component in space induced by $\phi$")
pl.ylabel("2nd component")
pl.subplot(2, 2, 4, aspect='equal')
pl.plot(X_back[:200, 0], X_back[:200, 1], "ro")
pl.plot(X_back[200:, 0], X_back[200:, 1], "bo")
pl.title("Original space after inverse transform")
pl.xlabel("$x_1$")
pl.ylabel("$x_2$")
pl.subplots_adjust(0.02, 0.10, 0.98, 0.94, 0.04, 0.35)
pl.show()
